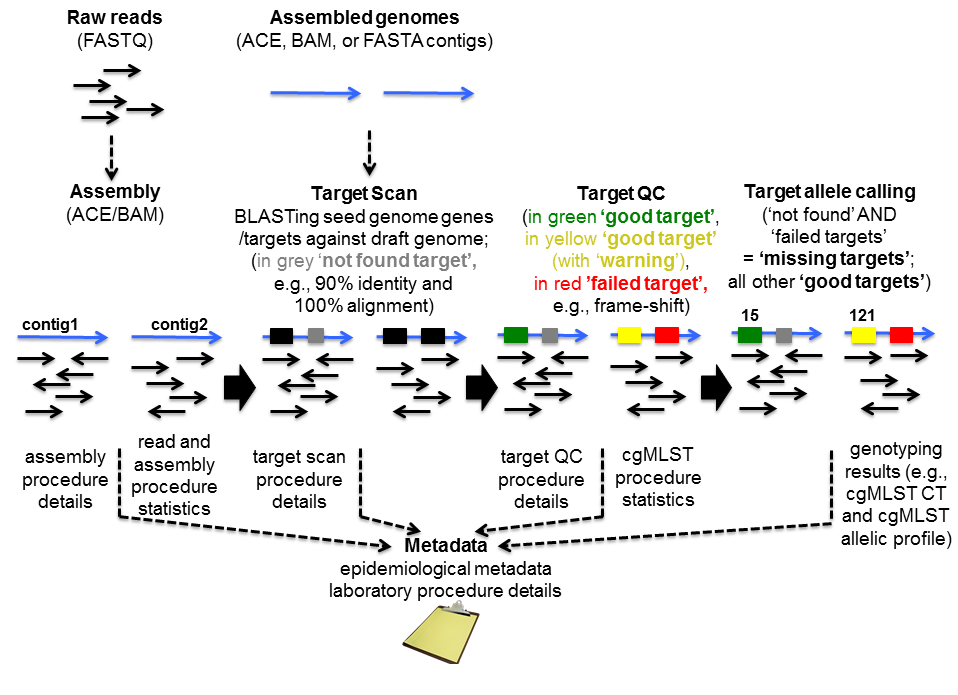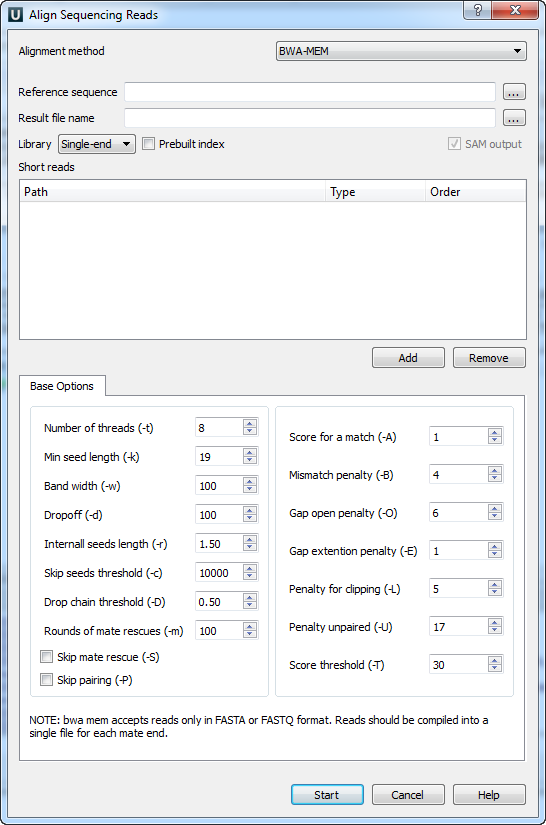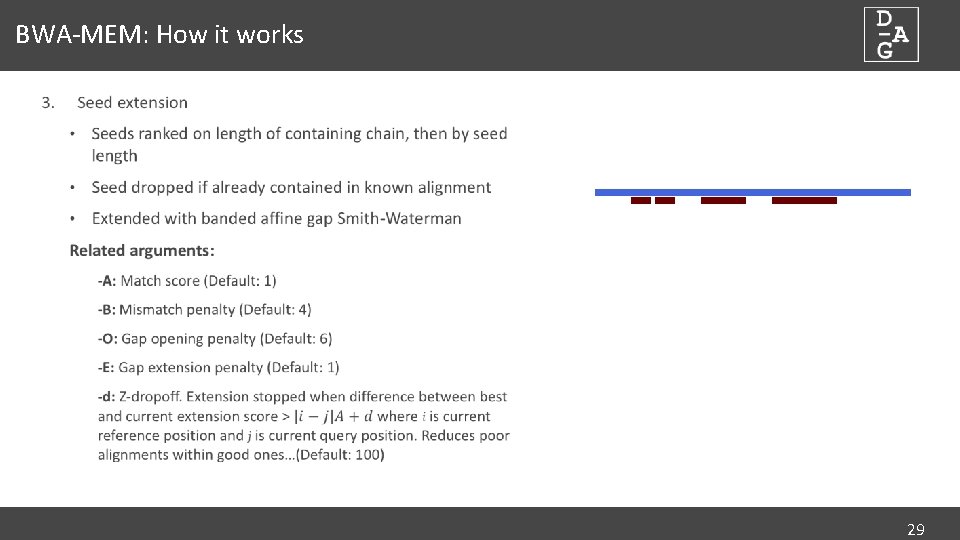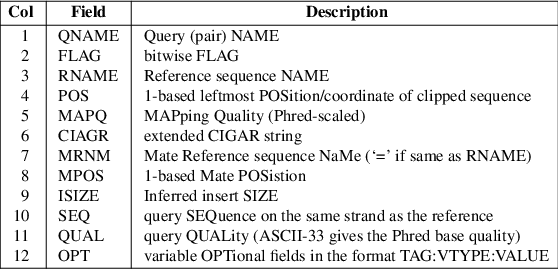
Pairwise alignment of nucleotide sequences using maximal exact matches | BMC Bioinformatics | Full Text
GASAL2: a GPU accelerated sequence alignment library for high-throughput NGS data | BMC Bioinformatics | Full Text
myExperiment - Workflows - cutadapt+BWA-MEM-mis1-i2,2-gapex2,2+filter_c5.0_c4.15+idxStats (Marc Ciosi) [Galaxy Workflow]