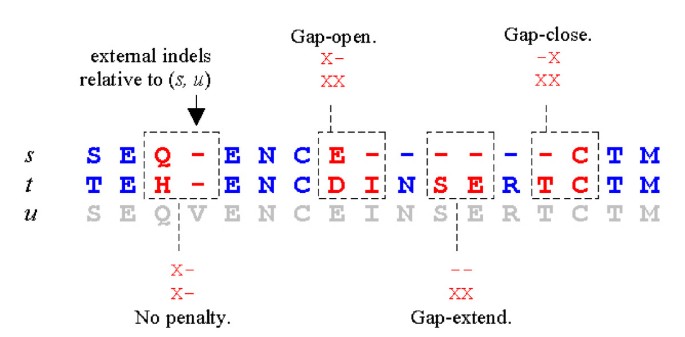
MUSCLE: a multiple sequence alignment method with reduced time and space complexity | BMC Bioinformatics | Full Text
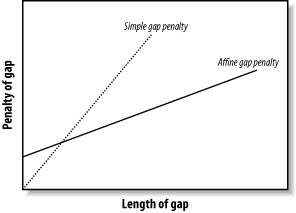
3.6 Variations :: Chapter 3. Sequence Alignment :: Part II: Theory :: Basic local alignment search tool (blast) :: Misc :: eTutorials.org
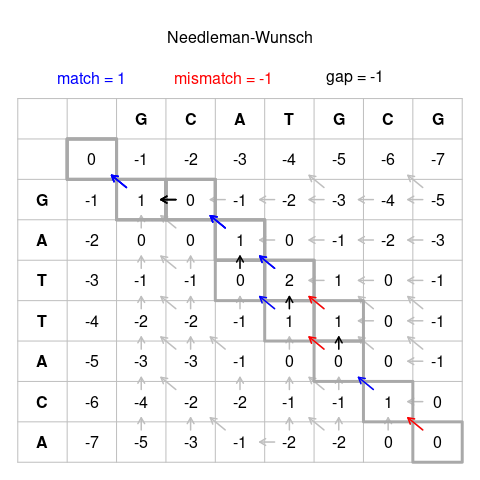
![If Score(i, j) denotes best score to aligning A[1 : i] and B[1 : j] Score(i-1, j) + galign A[i] with GAP Score(i, j-1) + galign B[j] with GAP Score(i, - If Score(i, j) denotes best score to aligning A[1 : i] and B[1 : j] Score(i-1, j) + galign A[i] with GAP Score(i, j-1) + galign B[j] with GAP Score(i, -](https://images.slideplayer.com/15/4857608/slides/slide_18.jpg)
If Score(i, j) denotes best score to aligning A[1 : i] and B[1 : j] Score(i-1, j) + galign A[i] with GAP Score(i, j-1) + galign B[j] with GAP Score(i, -
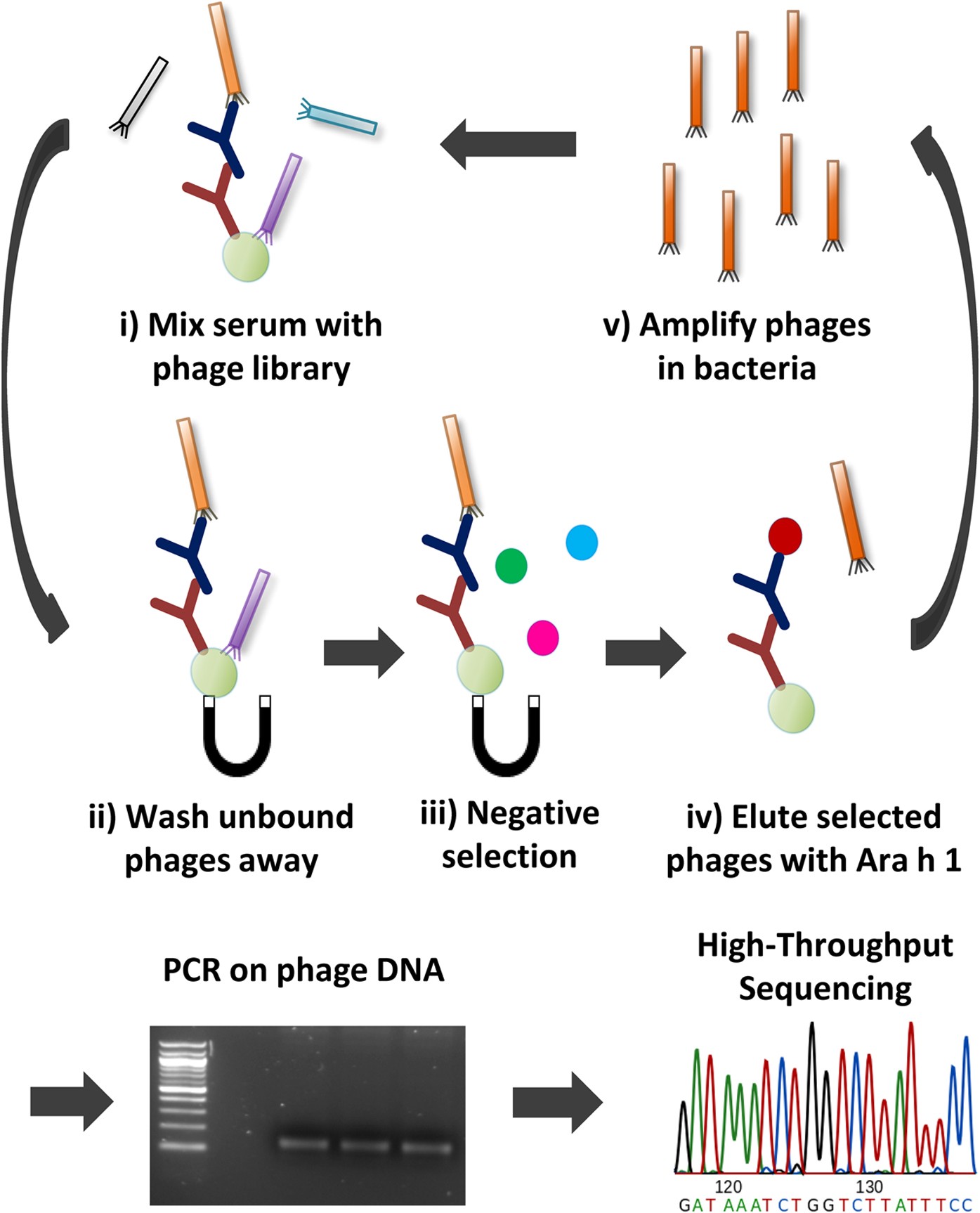
High-throughput sequencing enhanced phage display enables the identification of patient-specific epitope motifs in serum | Scientific Reports

AnchorWave: Sensitive alignment of genomes with high sequence diversity, extensive structural polymorphism, and whole-genome duplication | PNAS
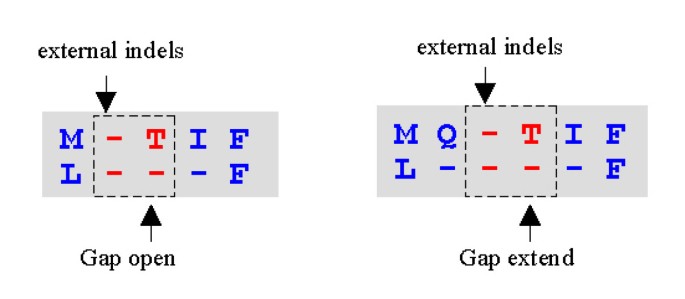
MUSCLE: a multiple sequence alignment method with reduced time and space complexity | BMC Bioinformatics | Full Text

AnchorWave: Sensitive alignment of genomes with high sequence diversity, extensive structural polymorphism, and whole-genome duplication | PNAS