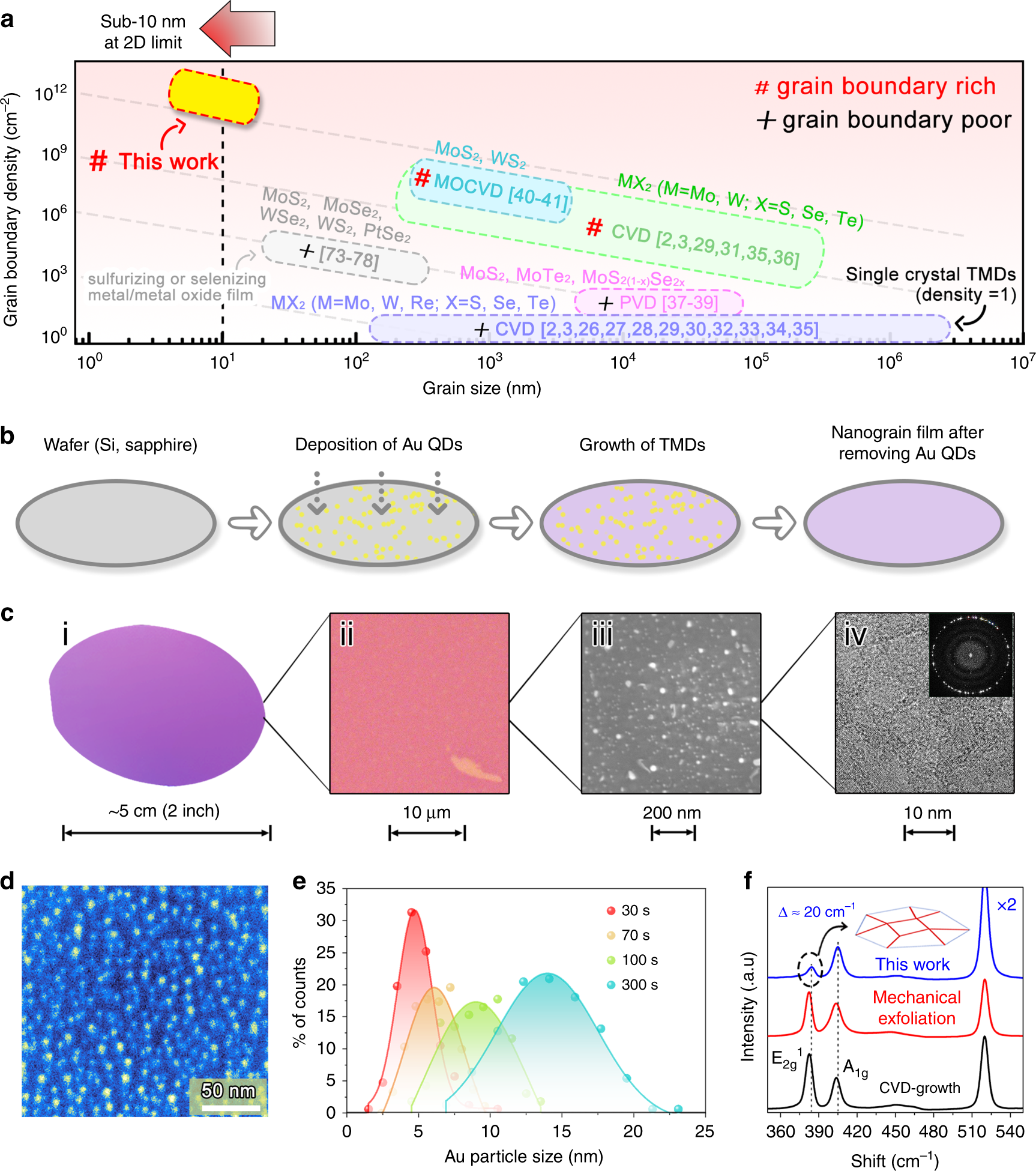
Frontiers | Genome Characterization of a Novel Wastewater Bacteroides fragilis Bacteriophage (vB_BfrS_23) and its Host GB124 | Microbiology
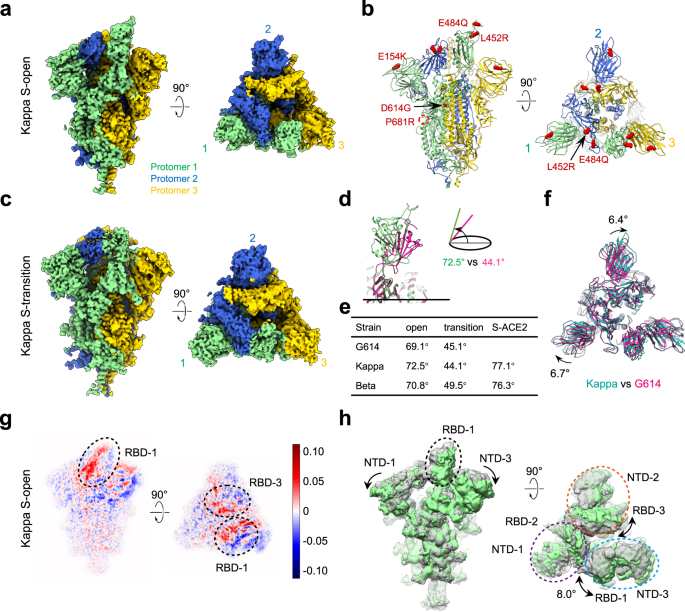
Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM | Nature Communications
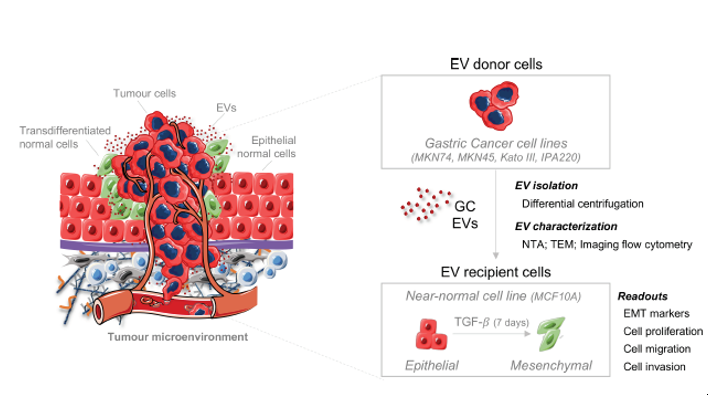
IJMS | Free Full-Text | Gastric Cancer Extracellular Vesicles Tune the Migration and Invasion of Epithelial and Mesenchymal Cells in a Histotype-Dependent Manner | HTML

Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery* - Journal of Biological Chemistry

Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM | Nature Communications

Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM | Nature Communications
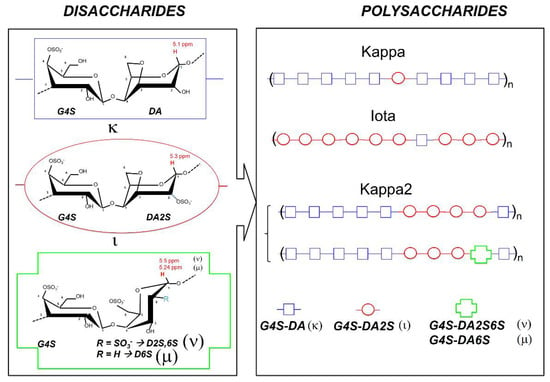
Polymers | Free Full-Text | Structure–Elastic Properties Relationships in Gelling Carrageenans | HTML
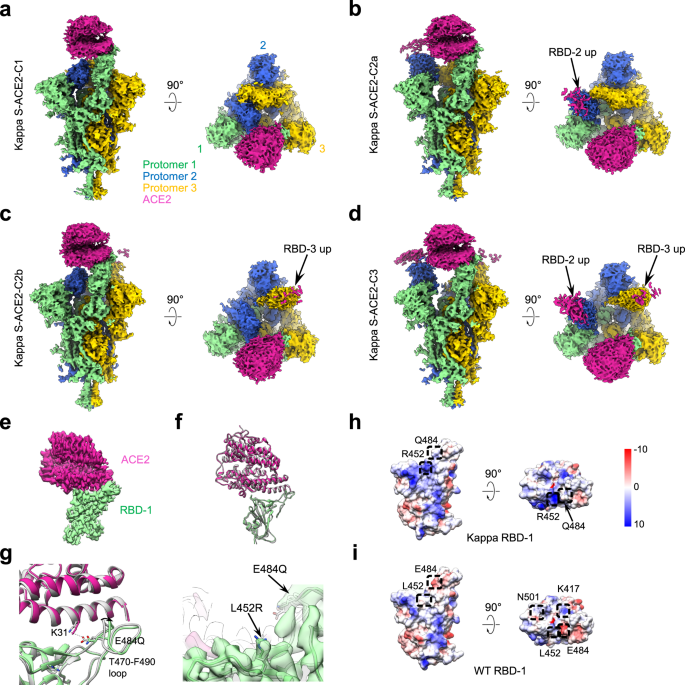
Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM | Nature Communications

In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies - ScienceDirect
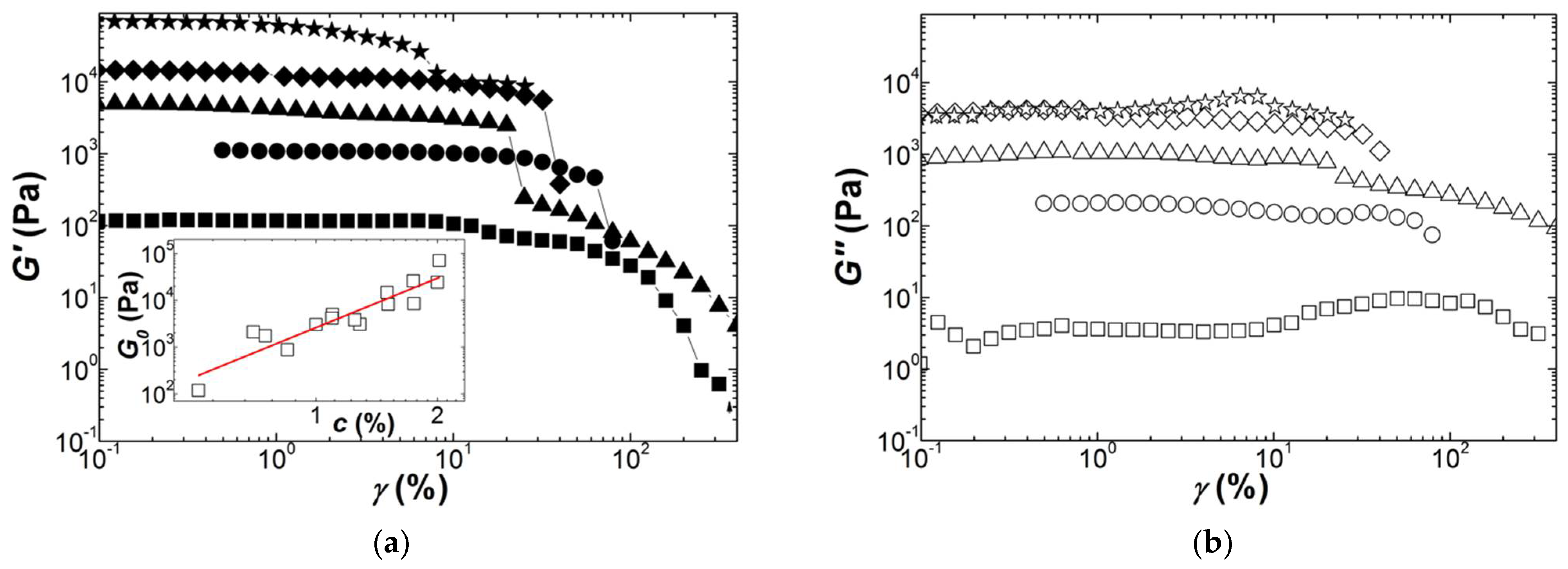
Polymers | Free Full-Text | Structure–Elastic Properties Relationships in Gelling Carrageenans | HTML

An interactive ImageJ plugin for semi-automated image denoising in electron microscopy | Nature Communications
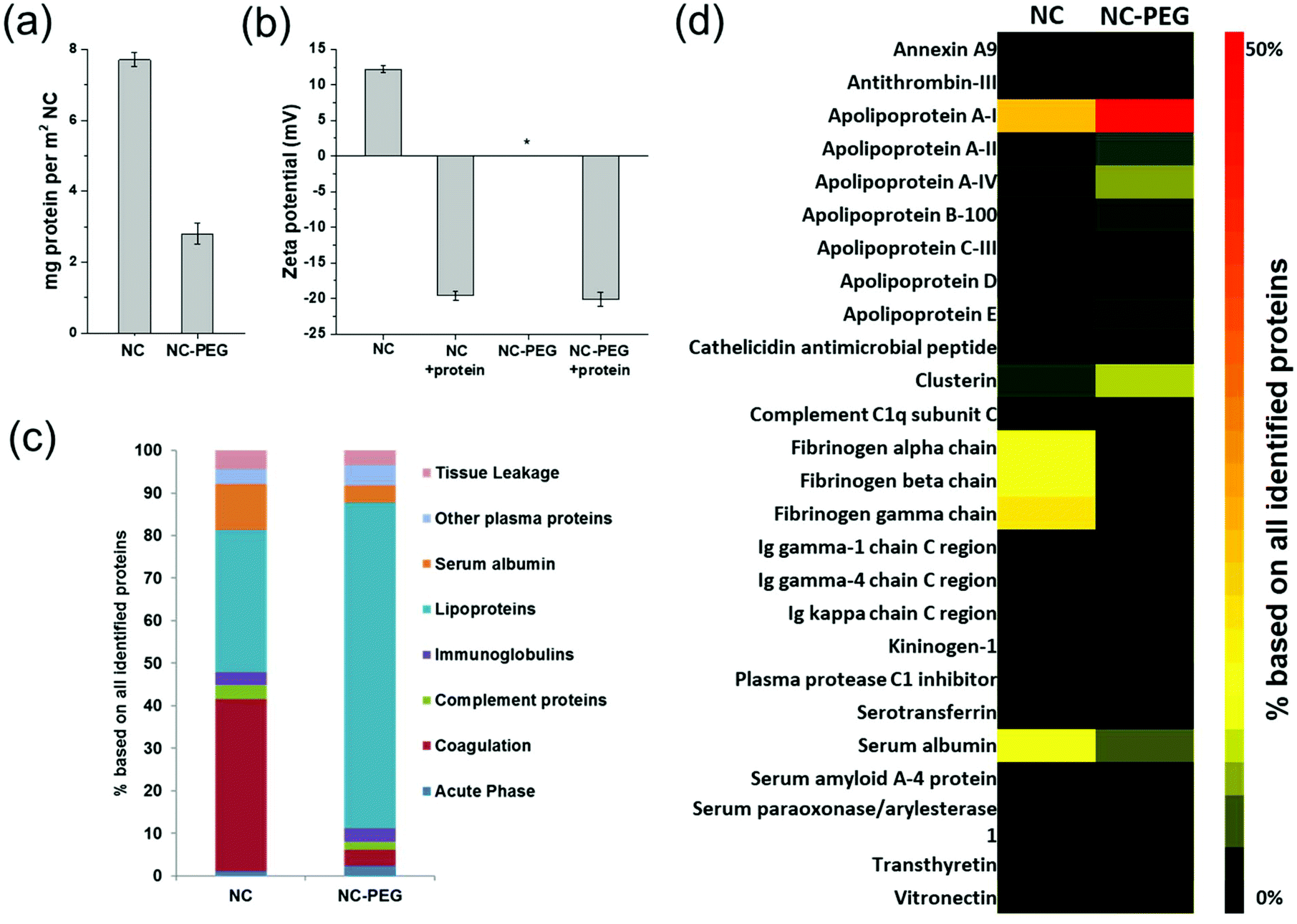
Controlling protein interactions in blood for effective liver immunosuppressive therapy by silica nanocapsules - Nanoscale (RSC Publishing) DOI:10.1039/C9NR09879H

Discovery of ultrapotent broadly neutralizing antibodies from SARS-CoV-2 elite neutralizers - ScienceDirect

Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD | bioRxiv

Cooperativity mediated by rationally selected combinations of human monoclonal antibodies targeting the henipavirus receptor binding protein - ScienceDirect

Paired heavy- and light-chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses - ScienceDirect

Ultrastructural insights into pathogen clearance by autophagy - Kishi‐Itakura - 2020 - Traffic - Wiley Online Library

turquoise nike dunks high tops shoes converse - Shirt Dark 313452 - Odlo Run Easy Men's Tank T - 15015
Sandfly Fever Sicilian Virus-Leishmania major co-infection modulates innate inflammatory response favoring myeloid cell infections and skin hyperinflammation

Influenza hemagglutinin-specific IgA Fc-effector functionality is restricted to stalk epitopes | PNAS