Activation of the HIV-1 Long Terminal Repeat by Nerve Growth Factor* - Journal of Biological Chemistry

Schematic of the HIV Long Terminal Repeat and Expected Products after... | Download Scientific Diagram

An endogenous retroviral long terminal repeat is the dominant promoter for human β1,3-galactosyltransferase 5 in the colon | PNAS

Long Terminal Repeat CRISPR-CAR-Coupled “Universal” T Cells Mediate Potent Anti-leukemic Effects: Molecular Therapy
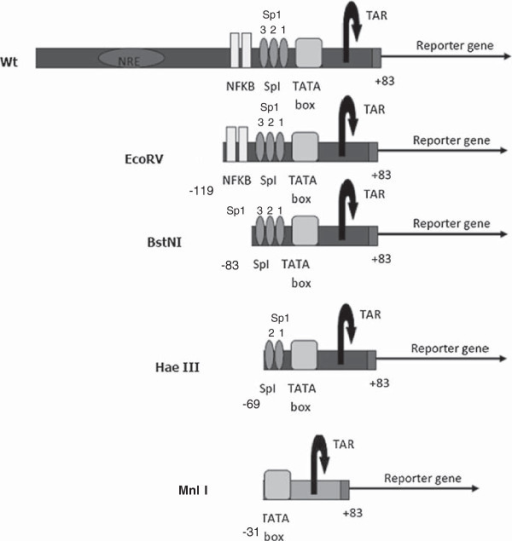
PLOS Pathogens: NFAT5 Regulates HIV-1 in Primary Monocytes via a Highly Conserved Long Terminal Repeat Site
PLOS Computational Biology: A machine learning based framework to identify and classify long terminal repeat retrotransposons
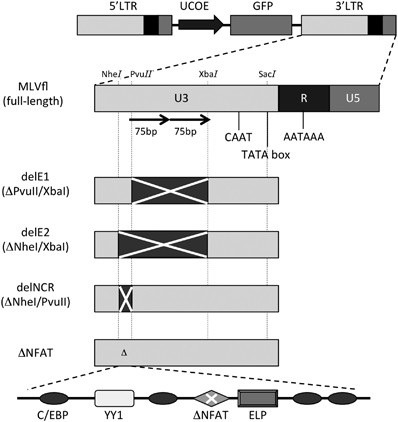
The long terminal repeat negative control region is a critical element for insertional oncogenesis after gene transfer into hematopoietic progenitors with Moloney murine leukemia viral vectors | Gene Therapy
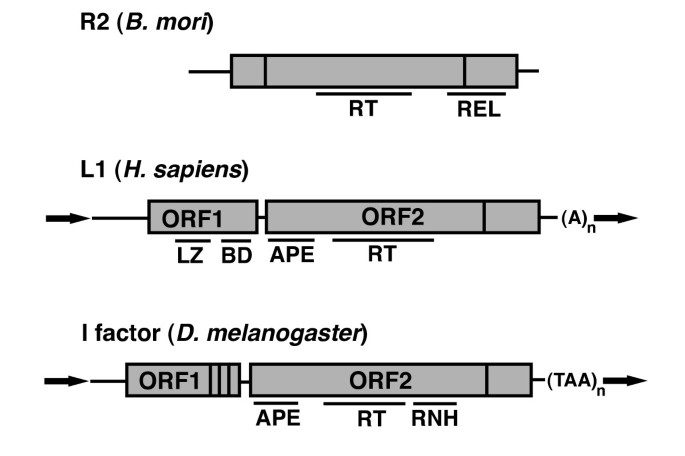
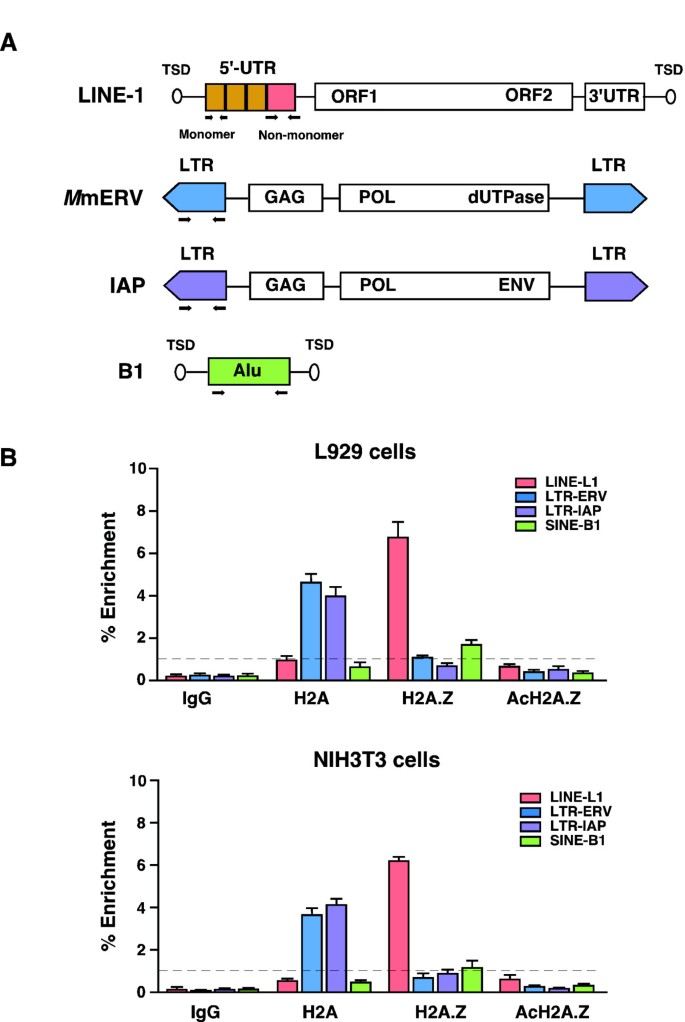
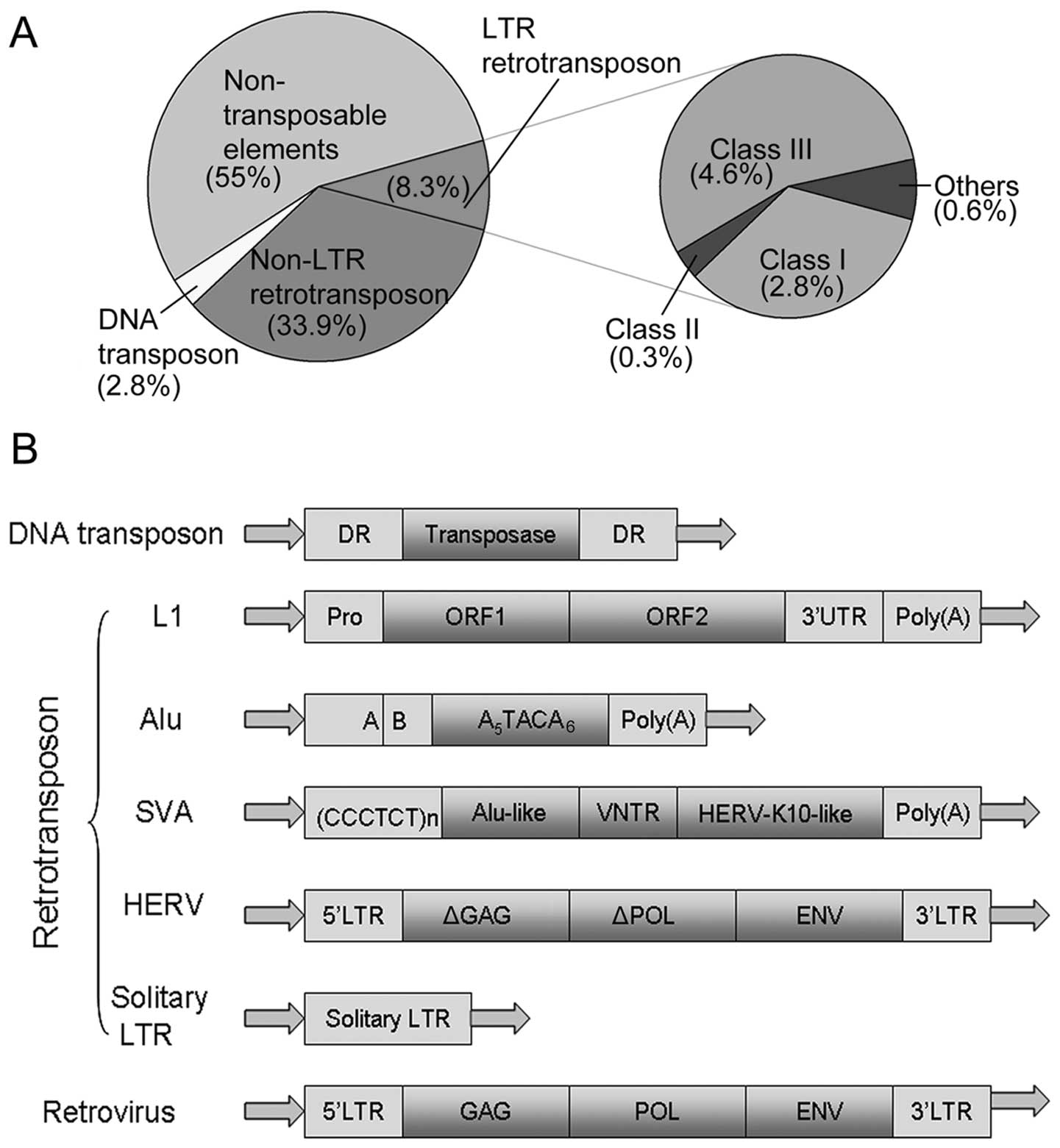
Non-long terminal repeat (non-LTR) retrotransposons: mechanisms, recent developments, and unanswered questions | Mobile DNA | Full Text

Changes to the HIV Long Terminal Repeat and to HIV Integrase Differentially Impact HIV Integrase Assembly, Activity, and the Binding of Strand Transfer Inhibitors* - Journal of Biological Chemistry

The Secondary Structure of the R Region of a Murine Leukemia Virus Is Important for Stimulation of Long Terminal Repeat-Driven Gene Expression | Journal of Virology

Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants | Nature Plants

LTR_retriever: a highly accurate and sensitive program for identification of LTR retrotransposons | bioRxiv